Introduction#
I am preparing a manuscript for submission to a scientific journal and decided to compose it in $\LaTeX$ using Overleaf. While Overleaf, an online $\LaTeX$ editor, has many collaboration features (including live, multi-person editing and review) many people, namely my PI, dislikes both $\LaTeX$ nor Overleaf, and instead prefers MS Word documents. One of Overleafs great feautres is the ability to use GitHub for version control. Therefore, I set up a GitHub Action workflow to test the compilation of the LaTeX document and convert it to docx Word format. It’s a free and easy way to satisfy my PI’s need for a Word docx and my desire to use $\LaTeX$ and Overleaf.
The Workflow#
Below is the entire GitHub Action workflow. To be used in any repository, it can be copied into a file with the path “.github/workflows/latex-action.yml” and the parameters can be customized, as discussed below.
name: Build LaTeX document
on: [push]
jobs:
build_latex:
runs-on: ubuntu-latest
steps:
- name: Set up Git repository
uses: actions/checkout@v1
- name: Compile LaTeX document
uses: xu-cheng/latex-action@master
with:
root_file: comutation-manuscript.tex
convert_via_pandoc:
runs-on: ubuntu-latest
steps:
- uses: actions/checkout@v2
- run: mkdir output
- uses: docker://pandoc/core:2.9
with:
args: >-
-s comutation-manuscript.tex
-f latex
-t docx
-o output/comutation-manuscript.docx
--bibliography reference_files/references.bib
--bibliography reference_files/R_citations.bib
--bibliography reference_files/additional_citations.bib
--csl nature-genetics.csl
- uses: actions/upload-artifact@master
with:
name: manuscript
path: output
Build $\LaTeX$#
The first “job” is build_latex.
This part of the workflow was taken from xu-cheng/latex-action on GitHub.
It uses his own Docker container to build and compile the $\LaTeX$ document.
To use this for your own project, just copy an past the above code snippet and change root_file: <your-latex-filename>.tex.
This job is not too complicated on our end and we are just looking for a “Success” or “Failure” output, so I will not go into further detail about how it works.
Convert to a Word Document#
The second job, convert_to_pandoc, converts the $\LaTeX$ file into a Word document of type docx.
I found the code for this portion at pandoc/pandoc-action-example and it uses a Docker container made available by Pandoc.
This job is a little more complicated so here are the steps it is taking:
- The standard GitHub
actions/checkout@v2is cloned - this is a standard step for most GitHub Actions so that the current git repository can be accessed by the rest of the Action. - A directory named “output” is created and will be where the final docx is stored.
- The pandoc Docker container is then used to set up the environment and the arguments for
pandocare submitted. Note that the>-is the YAML block chomping indicator so that the arguments can be placed on separate lines. The arguments used are:-s comutation-manuscript.tex: Thesflag stands forstandaloneand it takes the name of the $\LaTeX$ file.-f latex -t docx: “From” $\LaTeX$ “to” docx.-o output/comutation-manuscript.docx: The output file name; note that it will be put into the output directory made above.--bibliography reference_files/references.bib <etc>: The .bib citation files to use. Each use of the--bibliographyflag adds another file that pandoc will use.--csl nature-genetics.csl: The Citation Style Language (CSL) file with information on how to create the citations and bibliography. Others can be found at this GitHub repository or downloaded from Zotero.
- Finally, the output file can be saved to GitHub’s Artifacts and will be available for download.
Output#
If everything works as expected, then the Artifact should be available under the “Actions” tab of the repository and clicking on the most recent build. It should look something like the following screenshot.
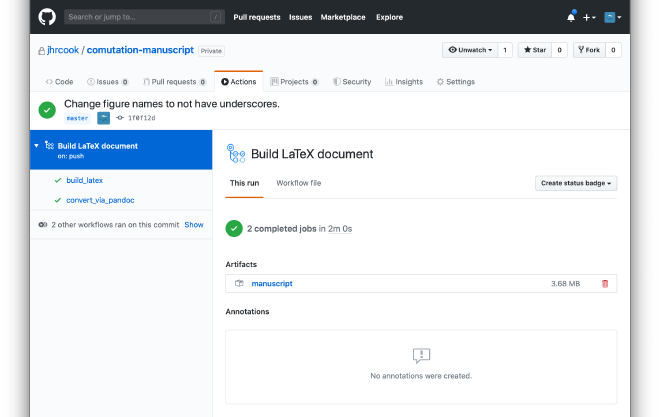
To keep an eye on the build process, I have selected the setting in the Notifications portion of the Settings on GitHub to send me an email on failure. I also have the dynamic badge on the README to show the current status of the Action’s build results. You can get the badge for your repository by clicking on the “Create status badge” in the “Action” tab (the button is visible in the above screenshot).

